
5 Tools for Ontology Alignment in Metadata
When working with metadata, ontology alignment tools are essential for connecting terms across different systems. For example, one database might use "cardiovascular disease", while another uses "heart condition." Without alignment, critical data remains disconnected. Here are five tools that help solve this problem:
- AgreementMakerLight (AML): Combines string similarity, linguistic analysis, and external knowledge for accurate alignment. Works well for large biomedical datasets and integrates easily with Java-based systems.
- LogMap: Uses reasoning and repair to align large, complex ontologies. It ensures consistency and is ideal for biomedical applications like SNOMED CT and NCIt.
- FCA-Map: Focuses on relationships between instances using Formal Concept Analysis. Best for cases where shared instance data exists.
- Ontology Xref Service (OxO): Aggregates existing mappings from trusted sources like UMLS and OLS. Ideal for quick lookups but doesn’t generate new mappings.
- BioPortal Mappings: Provides access to over 93 million pre-existing mappings across 1,246 ontologies. Useful for exploring connections between biomedical ontologies.
These tools vary in their approaches, scalability, and integration options. Below is a quick comparison for clarity:
| Tool | Alignment Approach | Best Use Case | Integration Options |
|---|---|---|---|
| AgreementMakerLight | Rules + lexical matching | Large-scale biomedical datasets | Java-based, CLI, GUI |
| LogMap | Logic-based + reasoning | Complex, large ontologies | Java, Web, CLI |
| FCA-Map | Instance-based (FCA) | Shared instance data scenarios | Limited automation |
| OxO | Mapping aggregation | Quick lookups of existing mappings | REST API, Docker |
| BioPortal Mappings | Pre-existing mappings | Accessing large mapping repositories | REST API, SPARQL, Web |
Choosing the right tool depends on your project's size, complexity, and technical requirements. Tools like AML and LogMap handle large-scale tasks effectively, while OxO and BioPortal simplify access to existing mappings.
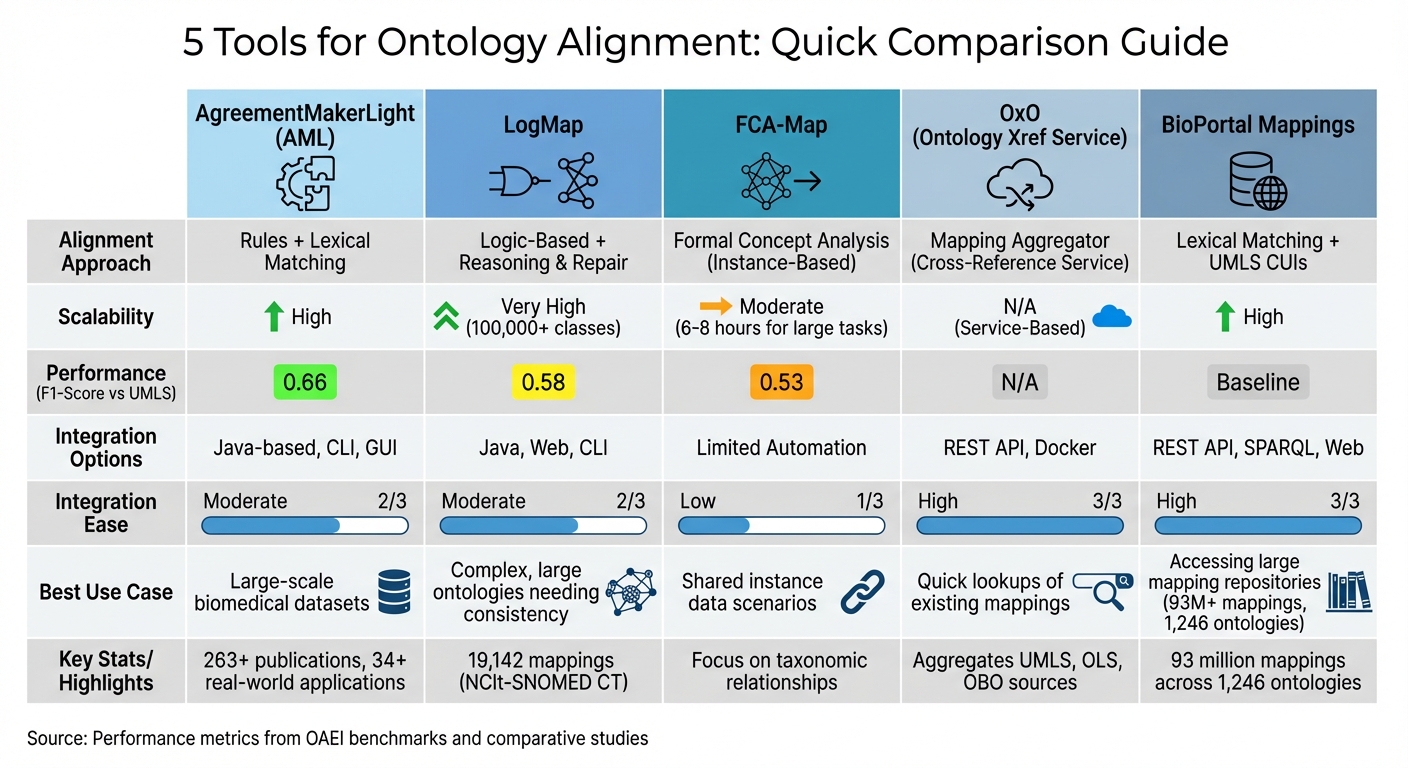
Comparison of 5 Ontology Alignment Tools: Features, Performance and Best Use Cases
Workshop Ontology Tools and Workflows
1. AgreementMakerLight
AgreementMakerLight (AML) is a powerful tool designed for aligning large biomedical ontologies. Developed by LASIGE at the University of Lisbon, it simplifies the process of ontology alignment while maintaining the adaptability needed for large-scale projects.
Alignment Approach
AML employs a combination of syntactic, linguistic, and semantic matching techniques to align ontologies effectively. It uses string similarity measures, linguistic analysis, and external domain knowledge to ensure accurate matches. This multi-layered method incorporates lexical matchers that rely on string comparisons and background knowledge from trusted databases. To make the process accessible, AML includes a user-friendly interface that simplifies alignment tasks, even for those without extensive technical expertise.
Supported Domains
Although AML can be applied across various fields, it particularly shines in biomedical metadata alignment. For example, researchers have used AML 2.0 to align datasets like NCIt, SNOMED CT, and ORDO, creating 6,463 mappings between ORDO and SNOMED CT and 18,887 mappings between NCIt and SNOMED CT. Its versatility has attracted users like NASA, Janssen Pharmaceutical Companies, and the Food and Agriculture Organization (FAO).
Performance Metrics
AML has consistently ranked among the top performers in the Ontology Alignment Evaluation Initiative (OAEI). It achieves median F-measures ranging from 1 to 2 and ranks within the top 3 to 4 in runtime performance. In the OAEI Anatomy track, Daniel Faria and his team highlighted AML's superior F-measure performance. In tests involving rare disease data, AML 2.0 achieved F1-scores of 0.66 when compared to UMLS and 0.55 against BioPortal alignments, outperforming competitors like LogMap 2.0 and FCA-Map. The tool has been referenced in over 263 independent publications and implemented in at least 34 documented real-world applications.
Ease of Integration with Metadata Systems
AML is designed for straightforward integration. It operates as a ready-to-use JAR file compatible with Java versions 1.7 through 1.9. Users can run it via command line for automated tasks or through its graphical interface for manual adjustments. It supports multiple ontology formats, including RDF/XML, OWL/XML, and Turtle, all of which are compatible with the OWL API. As an open-source project available on GitHub under the Apache-2.0 license, AML offers transparency and customization options for teams seeking to tailor their alignment workflows. This ease of use and adaptability make it a strong contender for exploring advanced ontology alignment solutions.
2. LogMap
LogMap is a logic-based ontology alignment tool specifically designed for the biomedical and life sciences sectors. Created by the Knowledge Representation and Reasoning Group at the University of Oxford, it is tailored to handle large, semantically complex ontologies such as SNOMED CT and the National Cancer Institute (NCI) thesaurus.
Alignment Approach
LogMap employs a mix of techniques to generate precise mappings. It begins with lexical indexing, using entity URIs and rdfs:label annotations to identify potential matches. Its reasoning and diagnosis features are built-in, allowing the tool to detect and resolve inconsistencies. This ensures that the resulting integrated ontology is consistent and free from unsatisfiable classes. Ernesto Jiménez-Ruiz, the lead developer, explains:
LogMap is one of the few matching systems that can efficiently match semantically rich ontologies containing tens (and even hundreds) of thousands of classes.
This method forms the backbone of its success in aligning complex biomedical ontologies.
Supported Domains
LogMap performs exceptionally well in biomedical metadata alignment, working with ontologies like the Foundational Model of Anatomy (FMA), SNOMED CT, and various disease and phenotype ontologies. It is also used to flag potential inconsistencies in BioPortal mappings. For instance, during a task involving NCIt and SNOMED CT - each containing over 150,000 and 350,000 classes, respectively - LogMap produced 19,142 mappings. Its advanced alignment techniques contribute to its competitive edge in handling such large-scale datasets.
Performance Metrics
In tests involving rare disease ontologies, LogMap 2.0 achieved an F1-score of 0.58 against UMLS Metathesaurus reference alignments and 0.55 against BioPortal references. Although these scores were slightly lower than those of AML, LogMap demonstrated much faster processing speeds, completing alignments in far less time compared to tools like FCA-Map, which required 6–8 hours. Its ability to deliver a "clean" set of mappings, thanks to its repair algorithms, makes it especially valuable for maintaining high data quality.
Ease of Integration with Metadata Systems
LogMap is open-source under the LGPL 3.0 license and integrates easily into Java applications via the OWL API (version 4). It offers a command-line interface for automated workflows and a web-based front-end for users who prefer a graphical interface. By leveraging the lexicon provided by ontologies - specifically rdfs:label annotations and entity URIs - it simplifies implementation into existing metadata systems. Similar to AML, LogMap's open-source availability and flexible interfaces make it a practical choice for integrating with metadata systems.
3. FCA-Map
FCA-Map takes a unique approach to ontology alignment by leveraging Formal Concept Analysis (FCA), moving away from traditional lexical or logic-based methods. Instead of relying on class names or structural organization, it focuses on the relationships between instances and their associated classes.
Alignment Approach
FCA-Map uses an extensional comparison strategy, meaning it examines the actual instances tied to classes rather than their labels or hierarchical positions. By creating formal concept lattices from shared instance data, it identifies matching classes across different ontologies. This method works only when the ontologies have a common set of instances, as it depends entirely on this shared data. Unlike LogMap, which emphasizes logic-based reasoning and repairing inconsistencies, FCA-Map’s instance-based approach is tailored for specific integration scenarios where shared data is present.
Ease of Integration with Metadata Systems
FCA-Map is particularly effective in environments where systems share a unified dataset of instances. In such cases, it can accurately align ontologies by analyzing how each organizes these shared instances. However, if no shared instance data exists, FCA-Map’s ability to perform alignment is significantly limited, as its functionality hinges on this commonality.
sbb-itb-f7d34da
4. Ontology Xref Service (OxO)
The Ontology Xref Service (OxO), developed by EMBL-EBI, acts as a mapping aggregator rather than a tool for generating new mappings. Its primary role is to collect and organize existing cross-references from trusted sources within the biomedical field.
Alignment Approach
OxO serves as a centralized hub, gathering cross-references from key repositories like the Ontology Lookup Service (OLS), the Unified Medical Language System (UMLS), and OBO cross-references. Instead of creating new mappings, it makes existing ones searchable and readily available.
The service employs a graph traversal system with a mapping distance metric. A distance of 1 indicates a direct, explicitly stated mapping between terms, while distances of 2 or 3 signify indirect mappings discovered through multiple "hops" in the graph. For instance, the MONDO disease ontology includes over 16,000 mappings to UMLS terms, all accessible via OxO. However, EMBL-EBI advises users to approach these mappings with care:
OxO aims to provide simple and convenient access to cross-references, but is not a mapping prediction service, so always treat these xrefs with caution, especially if you are seeking true equivalence between two ontologies.
These mappings are designed to integrate seamlessly with metadata systems.
Supported Domains
OxO is tailored specifically for biomedical metadata, supporting widely used standards such as MeSH, ICD-10, MONDO, NCIt, OMIM, and the Human Disease Ontology (DOID). This focus facilitates tasks like pathology annotation and clinical metadata integration.
Ease of Integration with Metadata Systems
Like other tools in this space, OxO offers flexible integration options and supports multiple export formats. Users can access the service through a REST API for automated integration, a web interface for manual searches, or a Dockerized version for local deployment, all under the Apache License 2.0. To use OxO, metadata must already include ontology term IDs (CURIEs), which serve as inputs for mapping searches. Results can be exported in TSV, CSV, or JSON formats, making it straightforward to incorporate them into existing workflows.
For the most reliable results, set the mapping distance to 1; higher distances may produce less accurate mappings. If OxO cannot locate a mapping, users can search the Ontology Lookup Service directly or request a term update from the relevant ontology community.
5. BioPortal Mappings
BioPortal stands out by gathering and organizing existing mappings rather than generating new ones. This approach helps streamline metadata integration, making it easier to connect and share information across various biomedical domains.
As of December 2025, BioPortal hosts an impressive repository of over 93 million mappings spanning 1,246 biomedical ontologies. Unlike tools that create new mappings, BioPortal acts as a centralized hub where researchers can explore, access, and download pre-existing mappings between ontology classes.
Alignment Approach
BioPortal uses several techniques to build and maintain its vast collection of mappings. One key method is lexical matching, which identifies exact matches between terms. Another approach involves UMLS mapping, where Concept Unique Identifiers (CUIs) are used to connect terms across different ontologies. As noted in the NCBO BioPortal Help documentation:
The assignment of terms to CUI is intended to allow the creation of maps between terms in different ontologies.
Mappings are refreshed automatically every night. Users can also interact with the system by creating, viewing, or editing mappings through its interface. These mappings include semantics and directionality, as defined by the user. To broaden its capabilities, BioPortal incorporates hierarchical relationships, such as "is-a" for OWL ontologies and "part-of" for OBO ontologies.
Supported Domains
BioPortal primarily serves the biomedical and healthcare sectors, covering areas like clinical and translational science, cancer informatics, and proteomics. Beyond these, it also supports biodiversity standards and international agricultural research through collaborations with CGIAR. Ontologies are grouped into categories such as those from UMLS, the OBO Foundry, and the WHO Family of International Classifications, enhancing its ability to support a wide range of domains.
Ease of Integration with Metadata Systems
BioPortal provides flexible integration options, including a REST API (accessible with an API key), a SPARQL endpoint, and a user-friendly web interface. It supports widely used ontology formats like OWL, OBO, and SKOS. However, some hosted mappings have historically lacked key metadata - such as confidence scores and provenance details - making them less suitable for critical automated tasks. To address this, the Simple Standard for Sharing Ontology Mappings (SSSOM) has become the recommended standard. SSSOM offers mappings in TSV or RDF formats enriched with metadata for better usability.
For efficiency, when searching across multiple ontologies, the system limits results to the top 100 matches unless filters are applied to narrow down the search.
Tool Comparison Table
Ontology alignment tools come with varying approaches, scalability, performance levels, and ease of integration. Here's a side-by-side comparison of five tools that are often used in production environments for ontology alignment:
| Tool | Alignment Approach | Scalability | Performance (F1-UMLS) | Integration Ease | Best For |
|---|---|---|---|---|---|
| AgreementMakerLight | Rules and lexical matching | High | 0.66 | Moderate (Java-based) | General-purpose tasks requiring fast, efficient matching |
| LogMap | Logic-based with reasoning & repair | Very High (100,000+ classes) | 0.58 | Moderate (Java/Web/CLI) | Large-scale biomedical ontologies needing consistency |
| FCA-Map | Formal Concept Analysis | Moderate (6–8 hours for large tasks) | 0.53 | Low (Slower processing) | Structural validation with taxonomic relationships |
| OxO | Aggregator / Cross-reference service | N/A (Service-based) | N/A | High (REST API) | Quick lookups of existing mappings across sources |
| BioPortal Mappings | Simple lexical (LOOM) | High | Baseline | High (REST/SPARQL/Web) | Accessing pre-existing biomedical mappings |
Key Insights on Tool Performance
- AgreementMakerLight stands out with an F1-score of 0.66, offering a balance of speed, quality, and efficiency. It's a reliable choice for general-purpose tasks where quick and effective matching is essential.
- LogMap excels in handling large-scale ontologies, especially in the biomedical field, thanks to its reasoning and repair capabilities. It achieves an F1-score of 0.58 and can manage ontologies with over 100,000 classes .
- FCA-Map uses Formal Concept Analysis to focus on structural validation and taxonomic relationships. While its F1-score is 0.53, it requires 6–8 hours for larger tasks, making it less suited for time-sensitive applications.
Each tool offers strengths tailored to specific needs, from high-speed matching to large-scale consistency checks or structural analysis.
Conclusion
When deciding on the best ontology alignment tool, the choice should align with the specific needs of your project. For handling large ontologies, tools like LogMap are well-suited due to their optimization for scale. On the other hand, smaller-scale tasks may benefit from LLM-based aligners for their ability to provide nuanced semantic mappings. If logical consistency and semantic depth are priorities, LogMap offers built-in reasoning and repair features to reduce mapping errors and maintain integrity.
Your technical setup also plays a role in tool selection. For Python developers, modular frameworks that integrate seamlessly with modern AI technologies may be more appealing. However, Java-based tools might require different infrastructure, so it's important to assess compatibility with your development environment.
Another key factor to weigh is the balance between complexity and speed. Lightweight aligners are ideal for large-scale tasks that demand quick processing, while LLM-based approaches can deliver richer semantic insights, albeit with higher computational demands. To ensure reliability, test tools against standardized benchmarks like the OAEI. As noted by Hamed Babaei Giglou and his team at TIB:
Existing tools are limited in scalability, modularity, and ease of integration with recent AI advances
This highlights the importance of thorough testing to pinpoint which limitations are most critical for your specific use case.
Finally, consider the tool's maintenance and documentation. Outdated tools may lack the support needed for modern workflows, and ensuring compatibility with your infrastructure can help you avoid installation and dependency issues later on.
FAQs
How does OntoAligner compare to other ontology alignment tools?
OntoAligner is a Python-based toolkit crafted for ontology alignment, offering a blend of traditional and cutting-edge features. It combines methods like fuzzy matching with advanced AI techniques, including large language models and retrieval-augmented generation, making it a versatile tool for diverse alignment tasks.
Although direct comparisons with other tools aren't provided, OntoAligner brings some standout features to the table. These include customizable alignment algorithms, built-in evaluation tools, and export capabilities. Its modular framework also allows researchers to encode ontological concepts in multiple ways, providing a flexible solution for improving metadata interoperability.
What should I consider when selecting an ontology alignment tool?
Choosing an ontology alignment tool hinges on understanding your project's unique requirements and objectives. Begin by assessing the tool's capabilities, such as its performance with large datasets, support for advanced techniques like AI or machine learning, and the option to integrate custom algorithms. Tools with a modular structure can be especially useful, offering easier customization and the ability to scale as your project grows.
Another key factor is the availability of resources like documentation, tutorials, and active community support. These can make implementation smoother and help resolve issues more efficiently. For additional guidance, consider exploring platforms like the Ontology Alignment Evaluation Initiative (OAEI), which provides detailed evaluations of different tools, highlighting their strengths and limitations to aid in your decision-making process.
Can these tools work with metadata systems outside of biomedical fields?
While the article doesn't delve into connecting with non-biomedical metadata systems, there are tools like the OntoAligner toolkit that cater to a wider range of applications. OntoAligner, for instance, facilitates semantic interoperability across diverse knowledge systems. It's even been applied in areas like eCommerce, hinting at its versatility beyond strictly biomedical contexts.